Following on from last weeks post in which we loaded a graph using Bio2RDF, this week we’ll look at some of the issues in parsing the data and then querying the graph.
Subclass Relationships
When we queried the SPARQL endpoint, one of the queries used the rdfs:subClassOf predicate which lead to an interesting result when examining the graph.
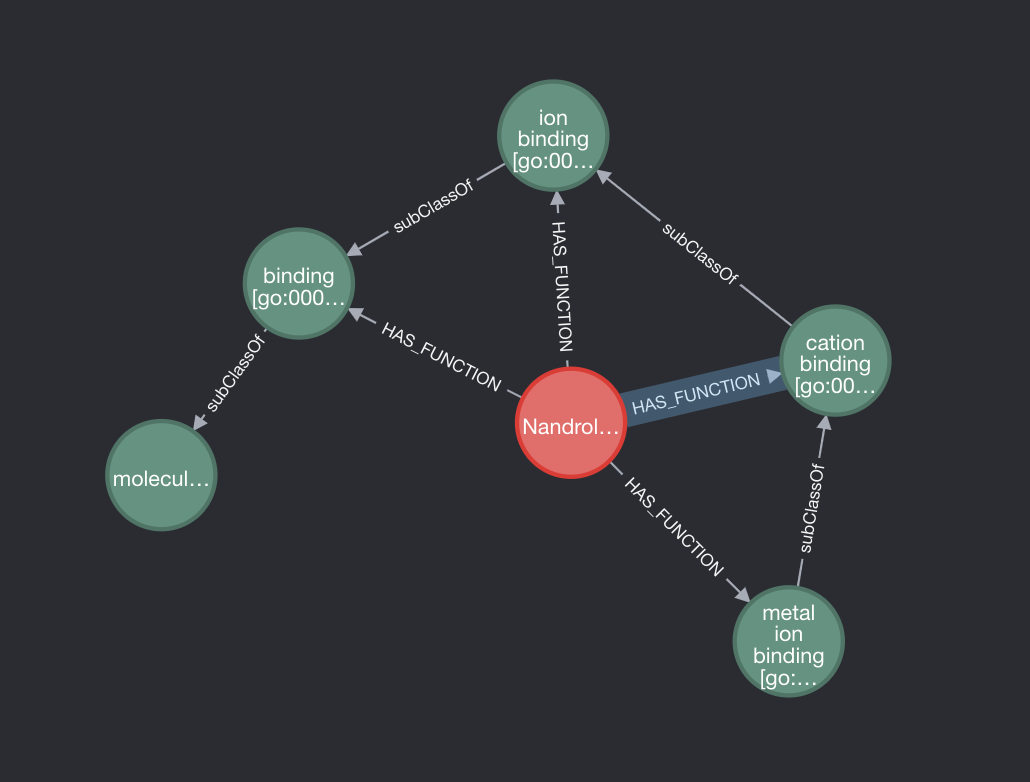
If we take Nandrolone as an example, we can see that it has multiple biological functions. However, many of these functions are parent classes of a lower function. It would make sense to delete these relationships as it should be a given that if Nandrolone has the function metal ion binding, which is subclassOf cation binding, then we can infer that it also has every parent function of it’s lowest level function. So how do we delete those relationships?
// Match every PED to its function
MATCH (p:PED)-[:HAS_FUNCTION]->(f:BioFunction)
// Find all the paths from these functions to their parents
MATCH path=(f)-[:subClassOf*]->(g)
// Ensure that f is the lowest level of the function hierarchy
WHERE NOT EXISTS (()-[:subClassOf]->(f))
WITH p,f,nodes(path) as nodes
// Find all the nodes in the path and delete the 'HAS_FUNCTION'
// relationship for all nodes except the first
UNWIND nodes as n
MATCH (p)-[rel:HAS_FUNCTION]->(n) WHERE id(n) <> id(f) DELETE rel
However, although the above query worked in most instances, it didn’t entirely solve our problem. If we run the same query as earlier for the biological functions of Nandrolone but also expand the lowest subclass, it turns out that function also has subclasses and therefore the query doesn’t fully eliminate all the redundant relationships.
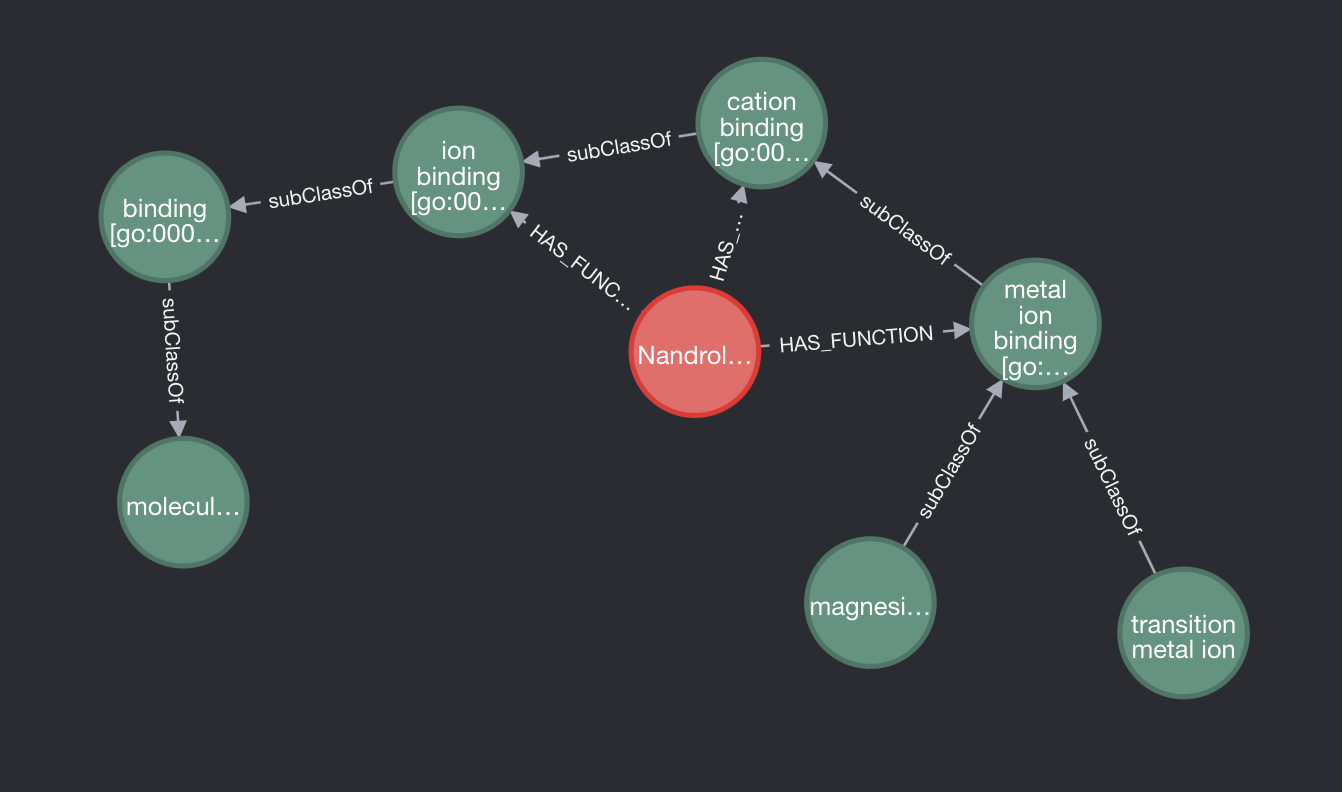
We can eliminate them using the following query.
MATCH (n:PED {name: 'Nandrolone'})-[:HAS_FUNCTION]->(f)-[:subClassOf*]->(bio:BioFunction)
WHERE EXISTS ((n)-[:HAS_FUNCTION]->(bio))
MATCH (n)-[rel:HAS_FUNCTION]->(bio)
DELETE rel
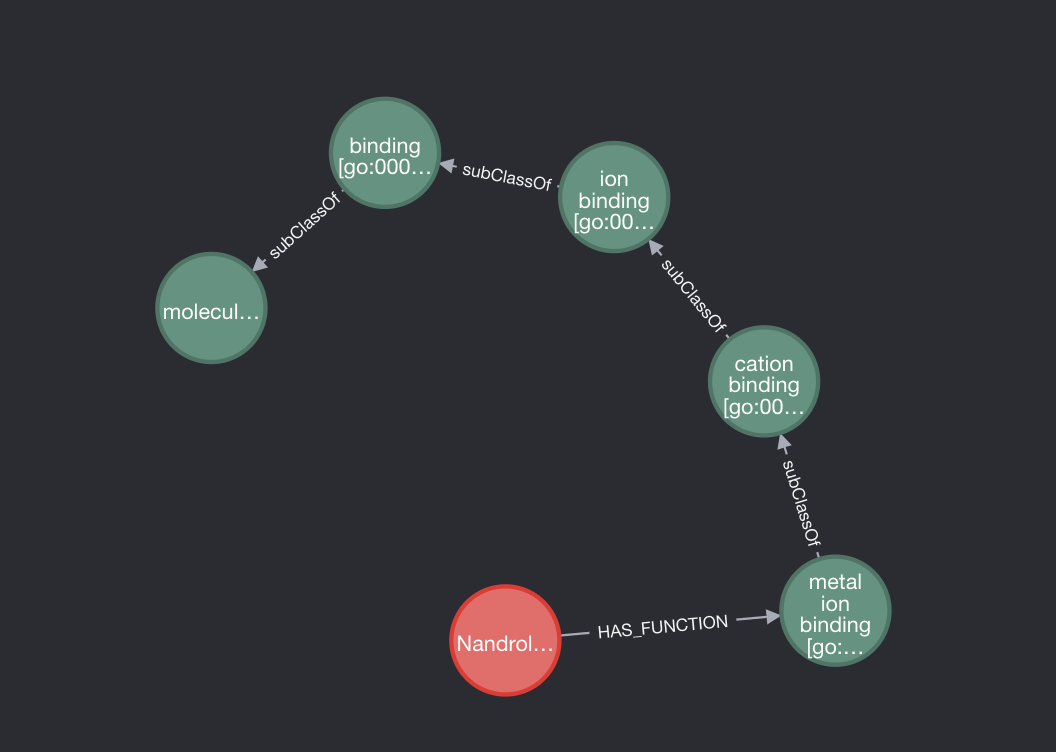
We can now see a much cleaner graph of subclasses for a PED.
Queries
Let’s look at some basic queries on the graph. Firstly we can look at how many biological functions each performance enhancing drug interacts with (caveat emptor - I’m not 100% sure how complete the Bio2RDF dataset is, so I’d advise caution when interpreting some of the query results).
MATCH (ped:PED)-[func:HAS_FUNCTION]-()
RETURN ped.name as drug , COUNT(func) as numFunctions
ORDER BY numFunctions DESC
+-------------------------------------+
| drug | numFunctions |
+-------------------------------------+
| "Metribolone" | 27 |
| "Androstenedione" | 17 |
| "Nandrolone" | 9 |
| "Clenbuterol" | 9 |
| "Zeranol" | 9 |
| "Methyltestosterone" | 8 |
| "Androstenediol" | 6 |
| "Danazol" | 6 |
| "Oxymetholone" | 5 |
+-------------------------------------+
We see Metribolone, Androstenedione and Nandrolone have the most biological functions that they interact with. Subsequently, each biological function involves interactions with many genes, so let’s see which genes are most prevelant.
MATCH (ped:PED)-[gene:INTERACTS_WITH]-() RETURN ped.name as drug , COUNT(gene) as numGenes ORDER BY numGenes
DESC
+---------------------------------+
| drug | numGenes |
+---------------------------------+
| "Metribolone" | 377 |
| "Androstenedione" | 48 |
| "Zeranol" | 33 |
| "Clenbuterol" | 24 |
| "Methyltestosterone" | 22 |
| "Danazol" | 19 |
| "Androstenediol" | 9 |
| "Oxymetholone" | 9 |
| "Nandrolone" | 4 |
+---------------------------------+
And conversely, we can also examine which genes interact with the largest number of performance enhancing drugs.
MATCH (g:Gene)<-[rel:INTERACTS_WITH]-(drug:PED)
RETURN g.name,COUNT(rel) as numDrugs ORDER BY COUNT(rel) DESC
+------------------------------------------------------------------------------------------------------------------+
| gene | numDrugs |
+------------------------------------------------------------------------------------------------------------------+
| "gene AR [ncbigene:367]" | 6 |
| "estrogen receptor 1 (symbol:ESR1, taxon:9606) [ncbigene:2099]" | 5 |
| "progesterone receptor (symbol:PGR, taxon:9606) [ncbigene:5241]" | 5 |
| "gene IL6 [ncbigene:3569]" | 4 |
| "cytochrome P450, family 19, subfamily A, polypeptide 1 (symbol:CYP19A1, taxon:9606) [ncbigene:1588]" | 3 |
| "gene LHB [ncbigene:3972]" | 3 |
| "follicle stimulating hormone, beta polypeptide (symbol:FSHB, taxon:9606) [ncbigene:2488]" | 3 |
| "kallikrein-related peptidase 3 [ncbigene:354]" | 3 |
| "nuclear receptor coactivator 1 (symbol:NCOA1, taxon:9606) [ncbigene:8648]" | 3 |
| "gene TNF [ncbigene:7124]" | 3 |
+------------------------------------------------------------------------------------------------------------------+
As expected when looking at genes that interact WADA’s banned anabolic agents, we see that the Androgen Receptor gene is involved with multiple PED’s along with estrogen and progesterone receptors.
Which genes are most influential?
I thought it would be interesting to see which genes are most influential in these various pathways. My first idea was to use a Cypher graph projection to create an OCCURS_WITH relationship for any two genes that are involved in the same pathway. Subsequently, I would pass this to the between centrality algorithm which detects the amount of influence a node has on the flow of information in a graph. Initially I tried this:
CALL gds.graph.create.cypher(
'geneGraphCypher',
'MATCH (g) WHERE g:Gene OR g:PED RETURN id(g) AS id',
'MATCH (g)<-[:INTERACTS_WITH]-(:PED)-[:INTERACTS_WITH]->(h) WHERE id(g) < id(h)
RETURN id(g) as source, id(h) as target'
)
YIELD graphName AS graph, nodeQuery, nodeCount AS nodes, relationshipCount AS rels
I now had a Cypher graph projection which I could feed into the betweeness centrality algorithm. Or so I thought. After trying to run this, and comparing the results to another method I’ll go over, I realised something wasn’t right. It turns out we can’t use a cypher projection in this instance because a cypher projection cannot use an undirected relationship. We need to create the relationships and then use those in a projected graph.
2.3. Relationship orientation The native projection supports specifying an orientation per relationship type. The Cypher projection will treat every relationship returned by the relationship query as if it was in NATURAL orientation. It is thus not possible to project graphs in UNDIRECTED or REVERSE orientation when Cypher projections are used. https://github.com/neo4j/graph-data-science/issues/113
Creating the relationships involved a fairly simple query.
MATCH (g:Gene)<-[:INTERACTS_WITH]-(ped:PED)-[:INTERACTS_WITH]->(g2:Gene)
WHERE id(g) < id(g2)
MERGE (g)-[:OCCURS_WITH]->(g2)
We can then create our graph projection specifying an undirected relationship.
CALL gds.graph.create(
'geneGraph',
'Gene',
{
OCCURS_WITH: {orientation: 'UNDIRECTED'}
}
);
And then subsequently call the graph data science between algorithm on the projected graph.
CALL gds.betweenness.stream('geneGraph')
YIELD nodeId, score RETURN gds.util.asNode(nodeId).name as geneName, score
ORDER BY score DESC LIMIT 5;
+------------------------------------------------------------------------------------------------------+
| geneName | score |
+------------------------------------------------------------------------------------------------------+
| "progesterone receptor (symbol:PGR, taxon:9606) [ncbigene:5241]" | 5762.870359203144 |
| "gene AR [ncbigene:367]" | 4460.623458469392 |
| "nuclear receptor coactivator 1 (symbol:NCOA1, taxon:9606) [ncbigene:8648]" | 3078.4971106238168 |
| "kallikrein-related peptidase 3 [ncbigene:354]" | 3078.4971106238168 |
| "gene IL6 [ncbigene:3569]" | 3030.9374285802037 |
+------------------------------------------------------------------------------------------------------+
It’s interesting to compare these to a more general aggregate which just looks at the number of cooccurences for a given gene.
MATCH (g:Gene)-[o:OCCURS_WITH]-()
RETURN g.name as geneName, count(o) as cooccurrences
ORDER BY COUNT(o) DESC LIMIT 5
+------------------------------------------------------------------------------------------------------------+
| geneName | cooccurrences |
+------------------------------------------------------------------------------------------------------------+
| "gene AR [ncbigene:367]" | 446 |
| "progesterone receptor (symbol:PGR, taxon:9606) [ncbigene:5241]" | 436 |
| "nuclear receptor coactivator 1 (symbol:NCOA1, taxon:9606) [ncbigene:8648]" | 431 |
| "kallikrein-related peptidase 3 [ncbigene:354]" | 431 |
| "gene LHB [ncbigene:3972]" | 426 |
+-----------------------------------------------------------------------------------------------------------+
The top four genes all occur in both query results although in slightly different order which must be related to the number of multiple hop cooccurrences I’d assume.
Closing Remarks
In a future post I’ll look to extend this to either more of WADA’s prohibited list or link the dataset up to another RDF store. Hopefully this has given a small insight into merging multiple sources of data and trying to create some insight. As always, any and all questions welcome!
