When exploring some linked datasets I stumbled across Bio2RDF, a resource for linked data for life sciences. This got me thinking how I could link this into WADA’s prohibited list and examine some of the genes linked to these Performance Enhancing Drugs (PED’s).
Scraping the Prohibited List
The first task was scraping the list of prohibited anabolic agents from the WADA website. This was a bit trickier than first thought as the website was dynamically loading the content when it was opened therefore using a simple requests.get wasn’t actually returning the content I wanted. As always, stackoverflow had an answer (which has actually turned out to be a revelation and could have saved me alot of time on some previous projects): if we use a webdriver like Selenium to open the page then call BeautifulSoup on the driver.page_source, we get the loaded version of the html.
import requests
from bs4 import BeautifulSoup
from selenium import webdriver
URL = 'https://www.wada-ama.org/en/content/what-is-prohibited/prohibited-at-all-times/peptide-hormones-growth-factors-related-substances-and-mimetics'
# Load the webpage
driver = webdriver.Firefox()
driver.get(URL)
# Retrieve the html content of the loaded page
html = driver.page_source
soup = BeautifulSoup(html)
# Find all list elements
data = soup.find_all('li')
# The returned list items also include some generic items which we can exclude
exclude_list = ['ADAMS','ADEL','Events','Newsroom', 'Contact Us', 'Q&A', 'English',
'Français','Other Languages', 'Who we are', 'What we do','What is Prohibited',
'Resources', 'All site', 'Substances & Methods', 'Resource', 'Home','Prohibited at All Times',
'Prohibited In-Competition','Prohibited in Particular Sports', 'English','Español',
'Français','Career Opportunities','Accessibility','Legal Statement','Privacy Policy','Site Map',
'Facebook','Instagram', 'Twitter', 'YouTube', 'LinkedIn']
# Clean the scraped list and append to the output list
output = []
for x in data:
if x.string is not None and x.string not in exclude_list:
output.append(x.string)
output= [x.split(' ')[0] for x in output]
>>> ['5-Androstenedione',
'7α-hydroxy-DHEA',
'7β-hydroxy-DHEA',
'7-Keto-DHEA',
'Androstenedione',
'Bolasterone',
'Boldenone',
'Boldione',
'Calusterone',
'Clostebol',
'Drostanolone',
'Epitestosterone',
'Fluoxymesterone',
'Formebolone',
'Gestrinone',
'Mestanolone',
'Mesterolone',
'Metandienone',
'Metenolone',
'Methandriol',
'Methylclostebol',
'Methyldienolone',
'Methyltestosterone',
'Metribolone',
'Mibolerone',
'Norboletone',
'Norethandrolone',
'Oxabolone',
'Oxandrolone',
'Oxymesterone',
'Oxymetholone',
'Quinbolone',
'Stanozolol',
'Stenbolone',
'Testosterone',
'Clenbuterol',
'Tibolone',
'Zeranol',
'Zilpaterol']
# The output isn't perfect but it will suffice for what we're trying to do
Querying Bio2RDF
The next step was creating a query to find these PED’s in the Bio2RDF resource, find any pathway associated with the drug and then any genes associated with these pathways. Firstly, we can create a Neo4j parameter to store the list of performance enhancing drugs.
:param PEDS => ['5-Androstenedione','7α-hydroxy-DHEA', '7β-hydroxy-DHEA','7-Keto-DHEA',
'Androstenedione', 'Bolasterone', 'Boldenone', 'Boldione', 'Calusterone', 'Clostebol',
'Drostanolone', 'Epitestosterone', 'Fluoxymesterone', 'Formebolone', 'Gestrinone',
'Mestanolone', 'Mesterolone', 'Metandienone', 'Metenolone', 'Methandriol',
'Methylclostebol', 'Methyldienolone', 'Methyltestosterone', 'Metribolone',
'Mibolerone', 'Norboletone', 'Norethandrolone', 'Oxabolone', 'Oxandrolone',
'Oxymesterone', 'Oxymetholone', 'Quinbolone', 'Stanozolol', 'Stenbolone',
'Testosterone', 'Clenbuterol', 'Tibolone', 'Zeranol', 'Zilpaterol']
The next step is to initiate our graph in the database.
call n10s.graphconfig.init({ handleVocabUris: "IGNORE"});
CREATE CONSTRAINT n10s_unique_uri ON (r:Resource)
ASSERT r.uri IS UNIQUE;
And finally we can pass our Bio2RDF query into the n10s procedure to retrieve our data from the database. We first take our list of performance enhancing drugs, unwind it and run the SPARQL query for each drug eventually returning all the nodes and relationships.
UNWIND $PEDS as ped
WITH 'PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX sio: <http://semanticscience.org/resource/>
PREFIX ctd: <http://bio2rdf.org/ctd_vocabulary:>
PREFIX bio_vocab: <http://bio2rdf.org/bio2rdf_vocabulary:>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX neo: <neo://voc#>
CONSTRUCT {
?drug a neo:PED ;
neo:name ?drugName ;
bio_vocab:identifier ?drugIdent .
?function a neo:BioFunction ;
neo:name ?functionLabel .
?classOfFunc a neo:BioFunction ;
neo:name ?classOfFuncLabel .
?drug neo:HAS_FUNCTION ?function.
?function neo:subClassOf ?classOfFunc .
?gene a neo:Gene ;
neo:name ?geneLabel .
?drug neo:INTERACTS_WITH ?gene .
} WHERE {
?drug a ctd:Chemical.
?drug dcterms:title ?drugName .
?drug dcterms:title "' + apoc.text.capitalize(ped) + '"@en .
?drug bio_vocab:identifier ?drugIdent .
OPTIONAL {
?drug ctd:has-function ?function .
?function rdfs:subClassOf ?classOfFunc .
?geneInteraction ctd:chemical ?drug .
?geneInteraction ctd:gene ?gene .
?gene rdfs:label ?geneLabel .
?function rdfs:label ?functionLabel .
?classOfFunc rdfs:label ?classOfFuncLabel } .
}
' as sparql
CALL n10s.rdf.import.fetch(
"https://bio2rdf.org/sparql?default-graph-uri=&query=" + apoc.text.urlencode(sparql),
"N-Triples",
{ headerParams: { Accept: "text/plain"}})
YIELD terminationStatus, triplesLoaded, triplesParsed, namespaces, extraInfo
RETURN terminationStatus, triplesLoaded, triplesParsed, namespaces, extraInfo
Visualising the Schema
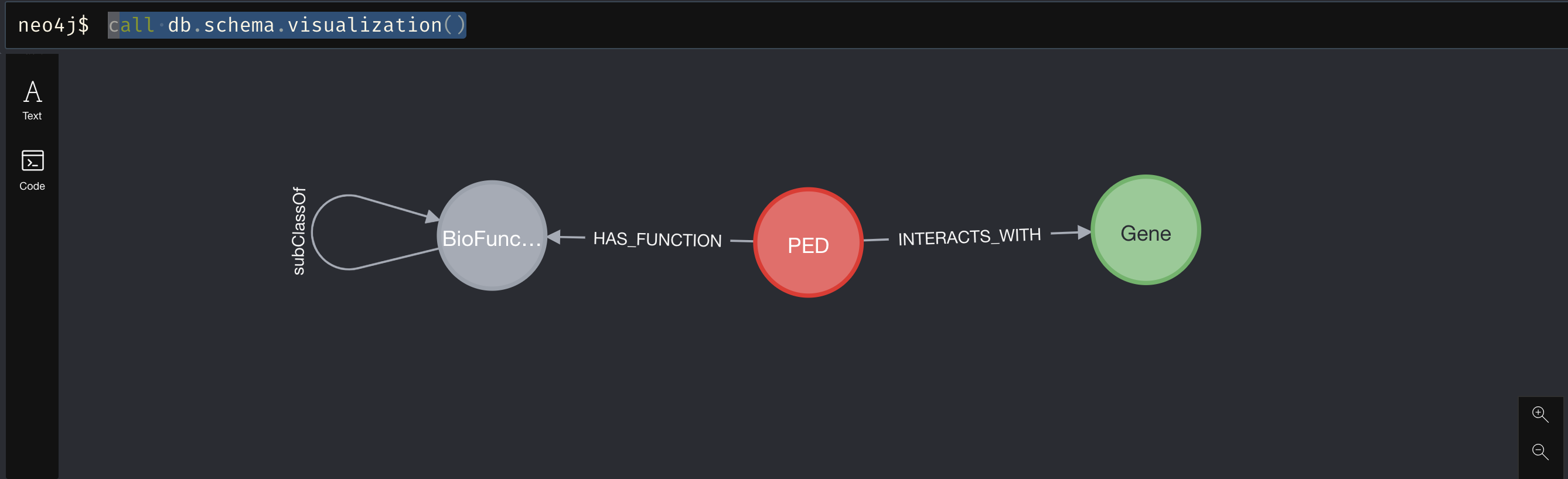
We can call db.schema.visualization() to see a representation of the schema.
To get an idea of the drug gene interactions we can also run a simple query to see how the different drugs may interact with similar genes and which genes are important in these pathways.
MATCH (g:Gene)<-[rel:INTERACTS_WITH]-(drug:PED) RETURN g, drug
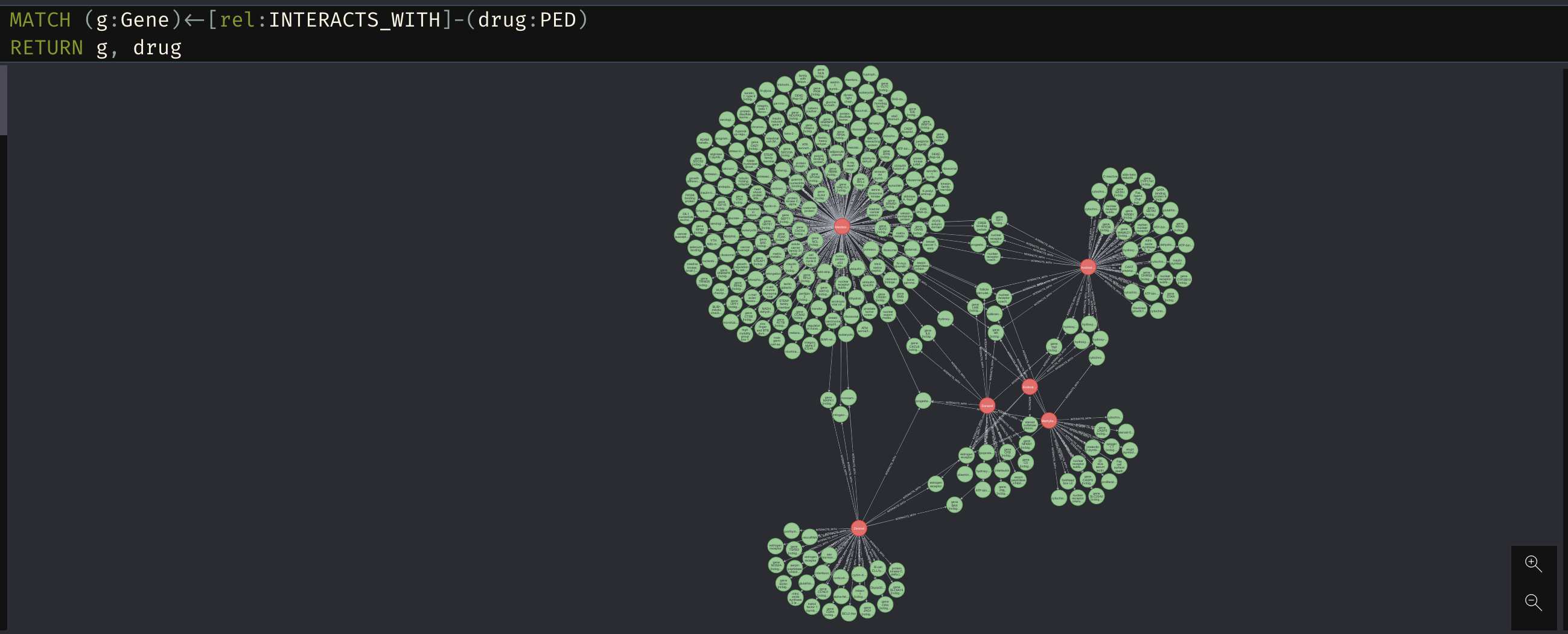
We can see from the screenshot above that there are clearly genes which interact with several of these performance enhancing drugs.
It’s a fairly short post this week but the main aim was just to go over how to query Bio2RDF with SPARQL. Next week’s post will look at this schema in more detail, look at how we can clean up some of the redundant relationships that the SPARQL query pulled in and finallly see what insight we can create from the data.
