I thought I’d have a look at a few different things this week as the title suggests: graph embeddings and named entity recognition through the medium of Sports Science research papers. So strap yourself in and let’s dive in…
Scraping The Data
The first stage was to collate some research papers from Pubmed. After having a search around it didn’t look like there was an API that would suit my needs so I ended up scraping the data myself.
The first step was to create some classes to store the data I wanted to scrape i.e. Authors and their research institutions along with details on academic papers including title, abstract, authors and it’s PubMed reference.
class Author:
def __init__(self, name, university):
self.name = name
self.university = university
class Paper:
def __init__(self, title, authors, abstract, pubmed_ref):
self.title = title
self.authors = authors
self.abstract = abstract
self.pubmed_ref = pubmed_ref
The next step was to have a rummage around the response html for a research papers page and figure out how to parse these various pieces of data. This is what I came up with…
paper_url = 'https://pubmed.ncbi.nlm.nih.gov'
def parse_paper(paper_ref):
# Extract the html for a given paper
r = requests.get(paper_url + paper_ref)
soup = BeautifulSoup(r.text)
# Get the paper title - always the main header H1
paper_title = soup.find('h1').text.strip()
print(paper_title)
# Not every paper had an abstract so make sure we catch this
# error if that's the case
try:
abstract = soup.find('div', {'class': 'abstract-content selected'}).text.strip()
except(TypeError, AttributeError) as e:
print(e)
abstract = None
# Similarly, not every paper had authors so catch this case
# if it occurs
try:
# Return the authors list from the html
authors_list = soup.find('div',{'class': 'authors-list'}).find_all('span')
# Create an empty list to store parsed Authors
authors_output = []
for authors in authors_list:
# Iterate through all authors for a paper
for author in authors.find_all('a', {'class':'full-name'}):
name = author['data-ga-label']
# The university labels weren't consistent so
# catch any errors here as well.
try:
uni = authors.find('a', {'class':'affiliation-link'})['title']
except(TypeError, AttributeError):
print('No university found')
uni = None
auth = Author(name, uni)
authors_output.append(auth)
except(TypeError,AttributeError) as e:
print(e)
authors_output = []
if abstract:
paper = Paper(paper_title, authors_output, abstract, paper_ref.replace('/',''))
else:
paper = Paper(paper_title, authors_output, None, paper_ref.replace('/',''))
return paper
Now we have a function to parse a single paper, we can then use this to get the information about multiple papers with the plan being to scrape all the papers from The Journal of Strength and Conditioning Research. A quick search on pubmed shows that there are 748 pages of results for papers from JSCR, each page with 10 papers. From this, it was simply a case of iterating through each results page and parsing every paper based on the pubmed reference using our parse_paper function.
papers = []
# Iterate through all results pages
for i in range(1,749):
URL = f'https://pubmed.ncbi.nlm.nih.gov/?term=%22Journal%20of%20strength%20and%20conditioning%20research%22%5BJournal%5D&sort=&page={i}'
r = requests.get(URL)
soup = BeautifulSoup(r.text)
# Find all paper references on the page
links = soup.find_all('a', {'class':'docsum-title'})
for research_paper in links:
paper = parse_paper(research_paper['href'])
papers.append(paper)
And now we can see an example of the data stored in each paper:
print(f'Title: {papers[0].title}\n')
print(f'Abstract: {papers[0].abstract}\n')
print(f'Pubmed Reference: {papers[0].pubmed_ref}\n')
for author in papers[0].authors:
print(author.__dict__)
>>> Title: Survey of Eccentric-Based Strength and Conditioning Practices in Sport
Title: Survey of Eccentric-Based Strength and Conditioning Practices in Sport
>>> Abstract: McNeill, C, Beaven, CM, McMaster, DT, and Gill, N. Survey of eccentric-
based strength and conditioning practices in sport. J Strength Cond Res 34(10):
2769-2775, 2020-Eccentric-based training (ECC) has been shown to be an effective
training strategy...
>>> Pubmed Reference: 32796422
{'name': 'Conor McNeill', 'university': 'Te Huataki Waiora School of Health,
Adams Center for High Performance, The University of Waikato, Tauranga, NZ;
and.'}
{'name': 'Christopher Martyn Beaven', 'university': 'Te Huataki Waiora School of
Health, Adams Center for High Performance, The University of Waikato, Tauranga,
NZ; and.'}
{'name': 'Daniel T McMaster', 'university': 'Te Huataki Waiora School of Health,
Adams Center for High Performance, The University of Waikato, Tauranga, NZ;
and.'}
{'name': 'Nicholas Gill', 'university': 'Te Huataki Waiora School of Health,
Adams Center for High Performance, The University of Waikato, Tauranga, NZ;
and.'}
Loading the Data into Neo4j
The next stage was to load the data into the Neo4j database. We’ll start by creating the driver in python and creating some constraints in the database which will help with the efficiency of loading and merging the nodes.
from neo4j import GraphDatabase
uri = "bolt://localhost:7687"
driver = GraphDatabase.driver(uri, auth=("neo4j", "password"))
with driver.session() as session:
session.run("CREATE CONSTRAINT UniquePaper IF NOT EXISTS ON (p:Paper) ASSERT p.pubmedRef IS UNIQUE ")
session.run("CREATE CONSTRAINT UniqueAuthor IF NOT EXISTS ON (a:Author) ASSERT a.name IS UNIQUE ")
session.run("CREATE CONSTRAINT UniqueUni IF NOT EXISTS ON (u:University) ASSERT u.name IS UNIQUE ")
Cleaning the Data
After a quick initial load of the data, it became pretty clear that although the paper details and author names had been scraped without an issue, the university associations we’re scraped as a single string and not as multiple separate strings for each author. Here’s one example:
1Physical Effort Laboratory, Sports Center, Federal University of Santa Catarina, Florianópolis, Brazil; and 2Human Performance Laboratory, Faculty of Kinesiology, University of Calgary, Calgary, Alberta, Canada.
And there were multiple other weird and wonderful ways the numbers were hidden in the text. We had to turn to everybody’s best friend - Regex. Here’s what I ended up using to split the text:
'(^\d+ *(?=[\. A-Za-z]*)|; *\d|[; ]+and *\d)')
I’ll go through each part…
- The outer most brackets are used as a capturing group to group mutliple different tokens together
- The first token -
^\d+ *(?=[\. A-Za-z]*): ^\d+ *- Starts with (^) one or more digit(s) (\d+) followed by 0 or more spaces ` *`(?=[\. A-Za-z]*)- Positive lookahead - must be followed by 0 or more full stops, spaces or letters.- The second token (using an or operator
|) -; *\d- a semi-colon followed by 0 or more spaces followed by a digit - The last token -
[; ]+and *\d- 1 or more semi-colon or space followed immediately by the wordand, followed by 0 or more spaces and finaly followed by a digit.
This was enough to cover pretty much all the different variations in how the numbers were spread throughout the text and did a fairly good job of splitting up the lists of university strings. Pop the regex expression in here for a better explanation.

Loading the Graph - Part 2
Now we know how we can clean up some of the data it’s time to load the graph.
# starts a neo4j driver session
with driver.session() as session:
tx = session.begin_transaction()
i = 0
for paper in papers:
try:
title = paper.title
abstract = paper.abstract
paper_ref = paper.pubmed_ref
# We can use the __dict__ function of a class to create a map
# for each of the classes varibles - useful for passing to Neo4j
author_map = [author.__dict__ for author in paper.authors]
query = """
// Merge the paper into the db
MERGE (p:Paper {pubmedRef: $pubmedRef})
ON CREATE SET
p.abstract =
CASE WHEN $abstract IS NOT NULL THEN $abstract ELSE NULL END,
p.title = $title
// Unwind the list of authors associated with a paper
WITH p, $authorMap as authors
UNWIND authors as author
MERGE (a:Author {name: author.name})
MERGE (a)-[:AUTHORED]->(p)
WITH id(p) as paperId, a, author
// Split the university string into separate institutions and
// create a relationship between these institutions and the
// research paper
CALL apoc.do.when(
author.university IS NOT NULL,
"WITH author.university as uniList, paperId as paperId
UNWIND apoc.text.split(uniList, '(^\d+ *(?=[\. A-Za-z]*)|; *\d|[; ]+and *\d)') as uni
MATCH (p:Paper) WHERE id(p) = paperId
MERGE (u:University {name: uni})
MERGE (u)<-[:ASSOCIATED_INSTITUTION]-(p)",
"",
{paperId:paperId, author:author})
YIELD value
RETURN 1
"""
tx.run(
query,
{
'title': title,
'abstract': abstract,
'pubmedRef': paper_ref,
'authorMap': author_map
}
)
i += 1
if i == 100:
tx.commit()
print(j, "records processed")
i = 0
tx = session.begin_transaction()
except Exception as e:
print(e, paper)
continue
tx.commit()
print(j, "lines processed")
And there we have it, the data loaded into the graph for some analysis in part 2…
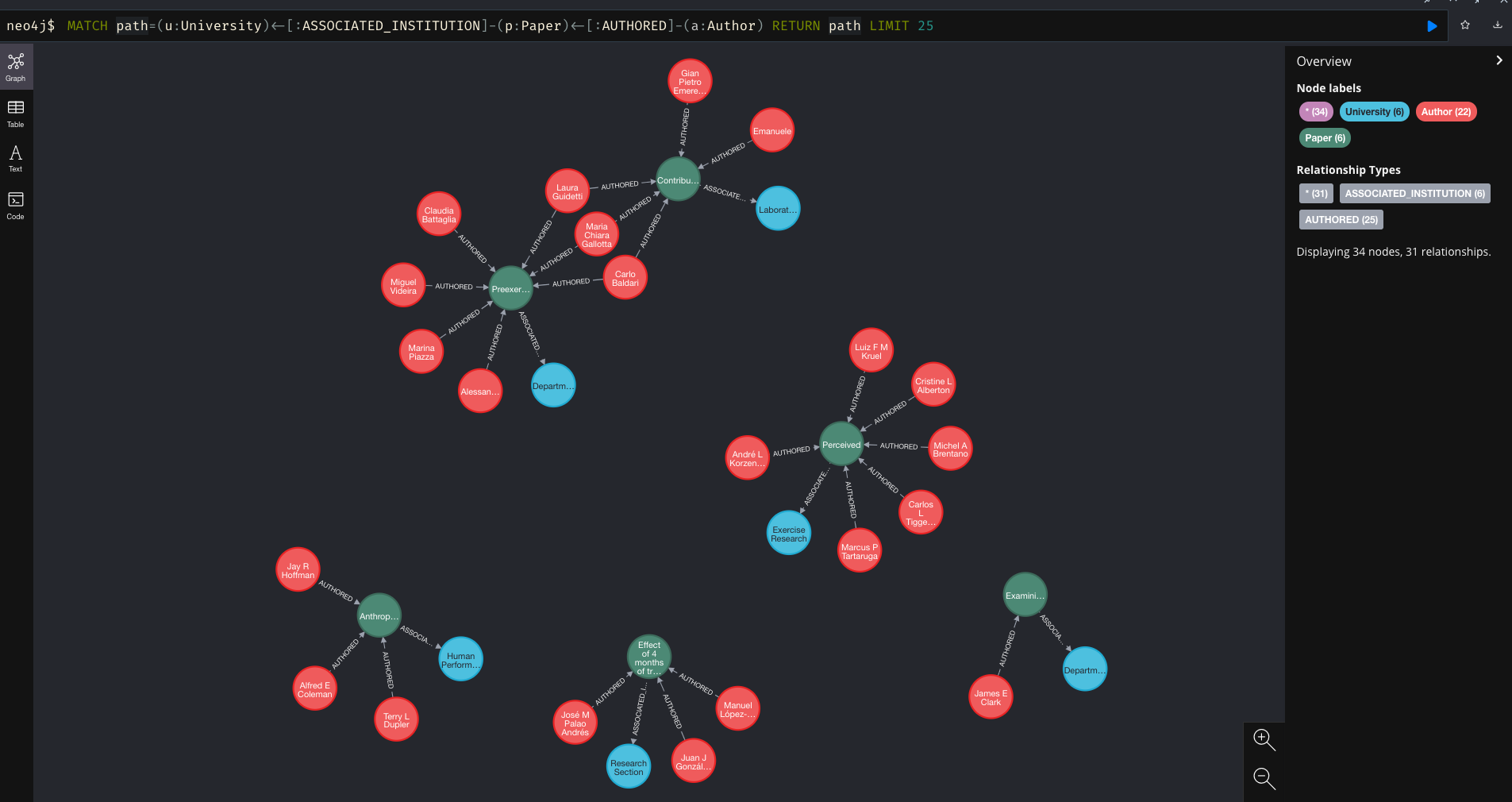
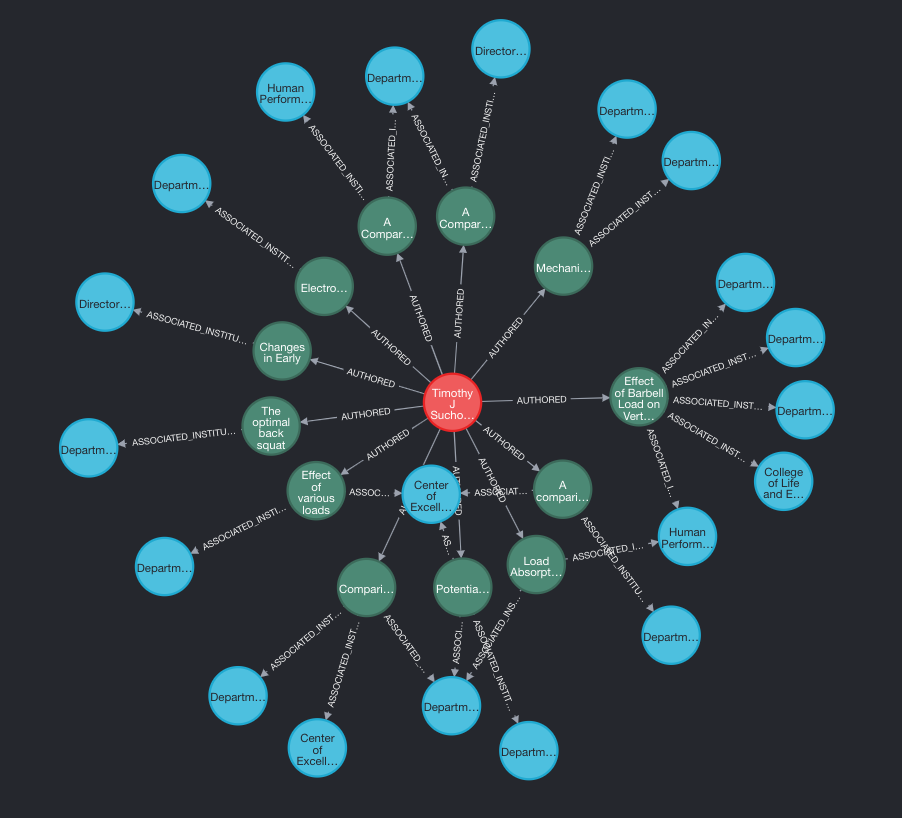
And finally, just to double check that my former colleague Michael ‘Fergy’ Ferguson’s all time favourite researcher is in the database, we can run a quick query…
MATCH path=(u:University)<-[:ASSOCIATED_INSTITUTION]-(p:Paper)<-[:AUTHORED]-(a:Author)
WHERE a.name CONTAINS 'Suchomel' RETURN path LIMIT 25
And we can see that Fergo will be very happy that he has a number of research papers within the newly created database.
Stay tuned for Part 2!
